RAG(Retrieval Augmented Generation)를 공부하면서 RAG pipeline을 직접 처음부터 구현해보고싶다는 생각을 하게 되었고, 적은 자원으로도 pipeline의 전 과정을 실행해볼 수 있는 오픈 소스를 발견하였다. 5GB+ VRAM을 가진 NVIDIA GPU만 있다면 local 환경에서도 실행할 수 있다.
- 링크: https://github.com/mrdbourke/simple-local-rag
- Setting: Python 3.11, CUDA 12.1, NVIDIA RTX 4090 + Windows 11
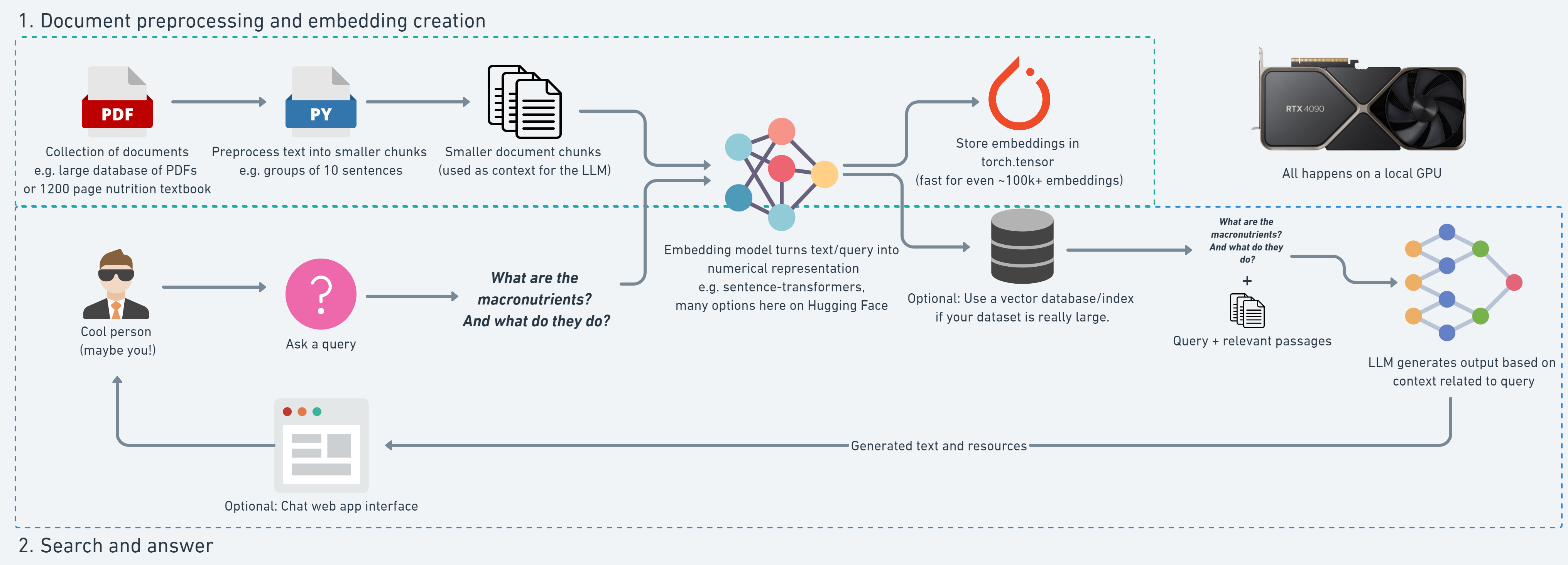
RAG 프로젝트 목표: NutriChat를 build하는 것
- 사람이 Nutrition Textbook 1200 페이지 분량의 PDF 버전에 대해 query 허용
- LLM이 textbook의 텍스트 지문들을 기반으로한 response 생성 후 반환
▶ RAG를 사용하여 hallucination을 방지하고, custom 데이터로 domain-specific 데이터에 대한 내용을 제공하자!
Document/Text Processing & Embedding Creation
준비 사항
- PDF 문서
- Embedding model
처리 과정
- PDF 문서 import하기
- Embedding 생성을 위해 Text 처리하기 (e.g. split into chunks of sentences).
- Embedding model로 Text Chunk의 Embedding 생성하기
- Embedding 저장하기
1. PDF 문서 import하기
1) open-source PDF textbook (Human Nutrition: 2020 Edition) 다운로드
# Download PDF file
import os
import requests
# Get PDF document
pdf_path = "human-nutrition-text.pdf"
# Download PDF if it doesn't already exist
if not os.path.exists(pdf_path):
print("File doesn't exist, downloading...")
# The URL of the PDF you want to download
url = "https://pressbooks.oer.hawaii.edu/humannutrition2/open/download?type=pdf"
# The local filename to save the downloaded file
filename = pdf_path
# Send a GET request to the URL
response = requests.get(url)
# Check if the request was successful
if response.status_code == 200:
# Open a file in binary write mode and save the content to it
with open(filename, "wb") as file:
file.write(response.content)
print(f"The file has been downloaded and saved as {filename}")
else:
print(f"Failed to download the file. Status code: {response.status_code}")
else:
print(f"File {pdf_path} exists.")2) PDF 페이지를 텍스트로 import (import PyMuPDF) → 각 페이지를 딕셔너리 형태로
import fitz
from tqdm.auto import tqdm # for progress bars, requires !pip install tqdm
def text_formatter(text: str) -> str:
"""Performs minor formatting on text."""
cleaned_text = text.replace("\n", " ").strip() # note: this might be different for each doc (best to experiment)
# Other potential text formatting functions can go here
return cleaned_text
# Open PDF and get lines/pages
# Note: this only focuses on text, rather than images/figures etc
def open_and_read_pdf(pdf_path: str) -> list[dict]:
"""
Parameters:
pdf_path (str): The file path to the PDF document to be opened and read.
Returns:
list[dict]: A list of dictionaries, each containing the page number
(adjusted), character count, word count, sentence count, token count, and the extracted text
for each page.
"""
doc = fitz.open(pdf_path) # open a document
pages_and_texts = []
for page_number, page in tqdm(enumerate(doc)): # iterate the document pages
text = page.get_text() # get plain text encoded as UTF-8
text = text_formatter(text)
pages_and_texts.append({"page_number": page_number - 41, # adjust page numbers since our PDF starts on page 42
"page_char_count": len(text),
"page_word_count": len(text.split(" ")),
"page_sentence_count_raw": len(text.split(". ")),
"page_token_count": len(text) / 4, # 1 token = ~4 chars, see: https://help.openai.com/en/articles/4936856-what-are-tokens-and-how-to-count-them
"text": text})
return pages_and_texts
pages_and_texts = open_and_read_pdf(pdf_path=pdf_path)
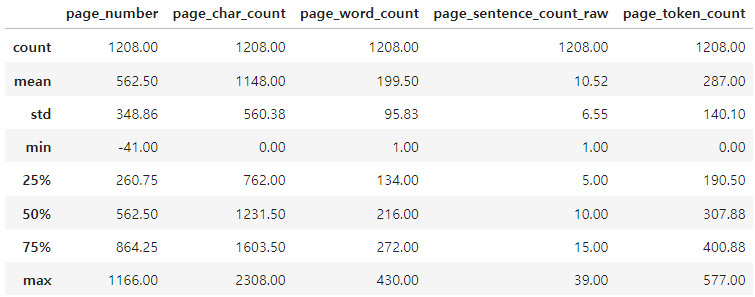
pages_and_texts[:2]pages_and_texts의 random sample
[{'page_number': -41, # adjust page numbers since our PDF starts on page 42
'page_char_count': 29,
'page_word_count': 4,
'page_sentence_count_raw': 1,
'page_token_count': 7.25, # 1 token = ~4 chars
'text': 'Human Nutrition: 2020 Edition'},
{'page_number': 84,
'page_char_count': 1372,
'page_word_count': 247,
'page_sentence_count_raw': 12,
'page_token_count': 343.0,
'text': '“Blood Flow Through the Heart” by OpenStax College / CC BY 3.0 The cardiovascular system is one of the eleven organ systems of the human body. Its main function is to transport nutrients to cells and wastes from cells (Figure 2.12 “Cardiovascular Transportation of Nutrients”). This system consists of the heart, blood, and blood vessels. The heart pumps the blood, and the blood is the transportation fluid. The transportation route to all tissues, a highly intricate blood-vessel network, comprises arteries, veins, and capillaries. Nutrients absorbed in the small intestine travel mainly to the liver through the hepatic portal vein. From the liver, nutrients travel upward through the inferior vena cava blood vessel to the heart. The heart forcefully pumps the nutrient-rich blood first to the lungs to pick up some oxygen and then to all other cells in the body. Arteries become smaller and smaller on their way to cells, so that by the time blood reaches a cell, the artery’s diameter is extremely small and the vessel is now called a capillary. The reduced diameter of the blood vessel substantially slows the speed of blood flow. This dramatic reduction in blood flow gives cells time to harvest the nutrients in blood and exchange metabolic wastes. Figure 2.11 The Blood Flow in the Cardiovascular System 84 | The Cardiovascular System']
3) 텍스트를 어떻게 split할지 파악하기 위해 EDA
- embedding model마다 다룰 수 있는 텍스트 size(input size of tokens)가 다름.
- sentence-transformers 중 all-mpnet-base-v2: 384 token 제한
- embedding text → 384 token들로 변형, 384 token이 넘는 텍스트는 자동으로 일부 정보를 잃고 384 token에 맞춰짐.
- sentence-transformers 중 all-mpnet-base-v2: 384 token 제한
2. Embedding 생성을 위해 Text 처리하기 (Chunking)
Ingest text -> Split it into groups/chunks -> Embed the groups/chunks -> Use the embeddings
- 규모가 큰 페이지들의 텍스트를 다루기 쉽고, 어떤 그룹의 문장들이 RAG 파이프라인에 도움이 되는지 알기 위해
- Options
- 간단한 규칙으로 split (text = text.split(". "))
- spaCy나 nltk같은 NLP library 사용
from spacy.lang.en import English
nlp = English()
# Add a sentencizer pipeline, see https://spacy.io/api/sentencizer/
nlp.add_pipe("sentencizer")
# Create a document instance as an example
doc = nlp("This is a sentence. This another sentence.")
assert len(list(doc.sents)) == 2
# Access the sentences of the document
list(doc.sents)
# 페이지들에 대한 sentencizing pipeline
for item in tqdm(pages_and_texts):
item["sentences"] = list(nlp(item["text"]).sents)
item["sentences"] = [str(sentence) for sentence in item["sentences"]]
# Count the sentences
item["page_sentence_count_spacy"] = len(item["sentences"])'sentences': ['consumed by the population, so that, on average, Americans obtain 44.6 percent of their foods in the meat and beans group from meats.',
' If you choose to follow a lacto-ovo vegetarian diet, the meats, poultry, and fish can be replaced by consuming a higher percentage of soy products, nuts, seeds, dry beans, and peas.',
'As an aside, the DGAC notes that these dietary patterns may not exactly align with the typical diet patterns of people in the United States.',
'However, they do say that they can be adapted as a guide to develop a more plant-based diet that does not significantly affect nutrient adequacy.',
' Table 6.6 Percentage of “Meat and Beans Group” Components in the USDA Base Diet, and Three Vegetarian Variations Food Category Base USDA (%) Plant-Based (%) Lacto-Ovo Vegetarian (%) Vegan (%) Meats 44.6 10.5 0 0 Poultry 27.9 8.0 0 0 Fish (high omega-3) 2.2 3.0 0 0 Fish (low omega-3) 7.1 10.0 0 0 Eggs 7.9 7.6 10.0 0 Soy products 0.9 15.0 30.0 25.0 Nuts and seeds 9.4 20.9 35.0 40.0 Dry beans and peas n/a* 25.0 25.0 35.0 Total 100.0 100.0 100.0 100.0 *The dry beans and peas are in the vegetable food group of the base diet.',
' Source: Vegetarian Food Patterns: Food Pattern Modeling Analysis.',
' US Department of Agriculture.',
' Appendix E-3.3.',
' http://www.cnpp.usda.gov/Publications/DietaryGuidelines/2010/ Proteins, Diet, and Personal Choices | 419'],
'page_sentence_count_spacy': 9}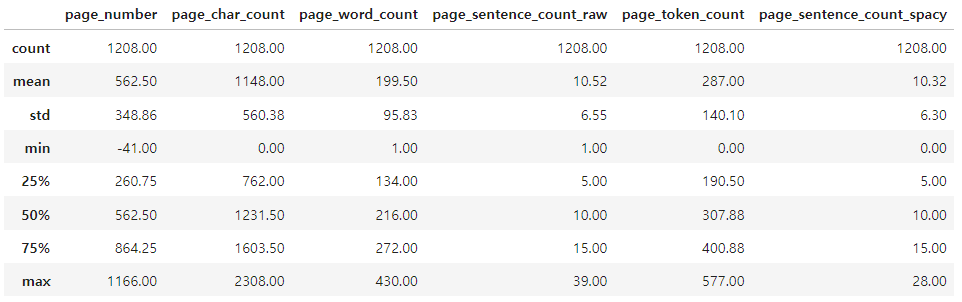
Chunking sentences
- 목적: embedding model이 수용 가능한 token 수를 넘지 않도록 하고, LLM이 가질 수 있는 token양인 context window가 제한되어 있음을 고려
- all-mpnet-base-v2 embedding model ≤ 384 tokens
- 문장 10개 (page_sentence_count) ⇒ ~287 tokens (page_token_count)
# 위에서 페이지별 문장 수의 평균 = 10
num_sentence_chunk_size = 10
# Create a function that recursively splits a list into desired sizes
def split_list(input_list: list,
slice_size: int) -> list[list[str]]:
"""
Splits the input_list into sublists of size slice_size (or as close as possible).
For example, a list of 17 sentences would be split into two lists of [[10], [7]]
"""
return [input_list[i:i + slice_size] for i in range(0, len(input_list), slice_size)]
# Loop through pages and texts and split sentences into chunks
for item in tqdm(pages_and_texts):
item["sentence_chunks"] = split_list(input_list=item["sentences"],
slice_size=num_sentence_chunk_size)
item["num_chunks"] = len(item["sentence_chunks"])
Splitting each chunk into its own item
문장의 chunk들 → Numerical representation으로 embedding
import re
pages_and_chunks = []
for item in tqdm(pages_and_texts):
for sentence_chunk in item["sentence_chunks"]:
chunk_dict = {}
chunk_dict["page_number"] = item["page_number"]
# Join the sentences together into a paragraph-like structure, aka a chunk (so they are a single string)
joined_sentence_chunk = "".join(sentence_chunk).replace(" ", " ").strip()
joined_sentence_chunk = re.sub(r'\.([A-Z])', r'. \1', joined_sentence_chunk) # ".A" -> ". A" for any full-stop/capital letter combo
chunk_dict["sentence_chunk"] = joined_sentence_chunk
# chunk에 대한 추가 정보를 알기 위해 dictionary 추가 (ex. relative info)
chunk_dict["chunk_char_count"] = len(joined_sentence_chunk)
chunk_dict["chunk_word_count"] = len([word for word in joined_sentence_chunk.split(" ")])
chunk_dict["chunk_token_count"] = len(joined_sentence_chunk) / 4 # 1 token = ~4 characters
pages_and_chunks.append(chunk_dict)
# How many chunks do we have?
len(pages_and_chunks) # 1843▶ 모든 textbook을 10개 이하의 문장들로 chunking!
BUT chunk_token 개수 편차가 크다는 문제점 발견!
▶ 30 token보다 작은 (3 chunk_tokens * 10개 chunk들) sample들 확인 ⇒ 페이지의 머리글이나 바닥글 ⇒ 제공하는 정보량이 적으므로 filtering
# Show random chunks with under 30 tokens in length
min_token_length = 30
for row in df[df["chunk_token_count"] <= min_token_length].sample(5).iterrows():
print(f'Chunk token count: {row[1]["chunk_token_count"]} | Text: {row[1]["sentence_chunk"]}')
Chunk token count: 5.25 | Text: Young Adulthood | 907
Chunk token count: 9.0 | Text: 1088 | Nutrition, Health and Disease
Chunk token count: 12.5 | Text: https://www.fda.gov/food/ 1022 | Food Preservation
Chunk token count: 19.25 | Text: 2018). Centers for Disease Control and 998 | The Causes of Food Contamination
Chunk token count: 12.5 | Text: Figure 11.2 The Structure of Hemoglobin Iron | 655
# filtering
pages_and_chunks_over_min_token_len = df[df["chunk_token_count"] > min_token_length].to_dict(orient="records")
pages_and_chunks_over_min_token_len[:2][{'page_number': -39,
'sentence_chunk': 'Human Nutrition: 2020 Edition UNIVERSITY OF HAWAI‘I AT MĀNOA FOOD SCIENCE AND HUMAN NUTRITION PROGRAM ALAN TITCHENAL, SKYLAR HARA, NOEMI ARCEO CAACBAY, WILLIAM MEINKE-LAU, YA-YUN YANG, MARIE KAINOA FIALKOWSKI REVILLA, JENNIFER DRAPER, GEMADY LANGFELDER, CHERYL GIBBY, CHYNA NICOLE CHUN, AND ALLISON CALABRESE',
'chunk_char_count': 308,
'chunk_word_count': 42,
'chunk_token_count': 77.0},
{'page_number': -38,
'sentence_chunk': 'Human Nutrition: 2020 Edition by University of Hawai‘i at Mānoa Food Science and Human Nutrition Program is licensed under a Creative Commons Attribution 4.0 International License, except where otherwise noted.',
'chunk_char_count': 210,
'chunk_word_count': 30,
'chunk_token_count': 52.5}]
3. Embedding model로 Text Chunk의 Embedding 생성하기
Text chunks → “learned” numerical representation (embedding vector)
▶ sentence-transformers library의 all-mpnet-base-v2 모델 사용
from sentence_transformers import SentenceTransformer
embedding_model = SentenceTransformer(model_name_or_path="all-mpnet-base-v2", device="cpu")
# 문장 list
sentences = [
"The Sentences Transformers library provides an easy and open-source way to create embeddings.",
"Sentences can be embedded one by one or as a list of strings.",
"Embeddings are one of the most powerful concepts in machine learning!",
"Learn to use embeddings well and you'll be well on your way to being an AI engineer."
]
# Sentences are encoded/embedded by calling model.encode()
embeddings = embedding_model.encode(sentences)
embeddings_dict = dict(zip(sentences, embeddings))
# 각 문장별 embedding 확인
for sentence, embedding in embeddings_dict.items():
print("Sentence:", sentence)
print("Embedding:", embedding)
print("")
# 단일 문장
single_sentence = "Yo! How cool are embeddings?"
single_embedding = embedding_model.encode(single_sentence)
print(f"Sentence: {single_sentence}")
print(f"Embedding:\n{single_embedding}")
print(f"Embedding size: {single_embedding.shape}")
Embedding size: (768,)
# text -> batched operations
text_chunk_embeddings = embedding_model.encode(text_chunks,
batch_size=32,
convert_to_tensor=True) # optional to return embeddings as tensor instead of array
4. Embedding 저장하기
pages_and_chunks_over_min_token_len 딕셔너리 리스트 → 저장
text_chunks_and_embeddings_df = pd.DataFrame(pages_and_chunks_over_min_token_len)
embeddings_df_save_path = "text_chunks_and_embeddings_df.csv"
text_chunks_and_embeddings_df.to_csv(embeddings_df_save_path, index=False)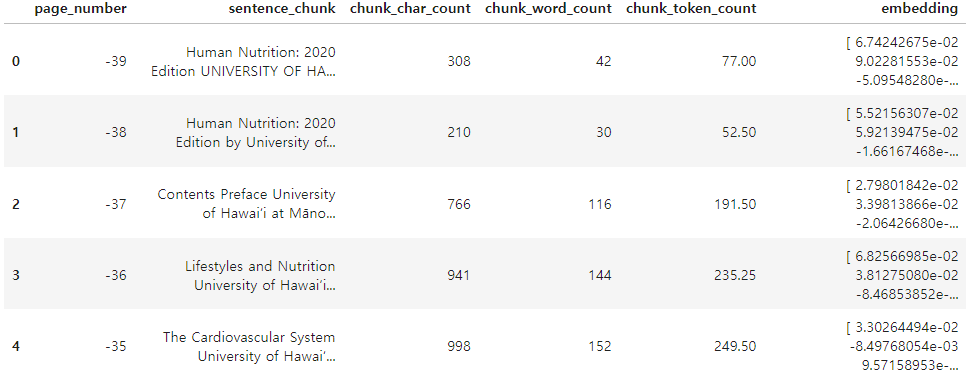
FAQ - Chunking & Embedding
- embedding model 선택
- Text chunking/splitting 방식
- pinecone → chunking
- langchain → splitting
- embedding 생성 시 고려해야할 것 (+ compute/storage)
- Size of input
- Size of embedding vector
- Size of model
- Open or closed
- embedding 저장 위치
- 약 10만 개 이하 → np.array 또는 torch.tensor
- 약 10만 개 이상 → vector database
'Research & Study Notes' 카테고리의 다른 글
| Comparison of Keyword Extraction Methods (0) | 2025.01.22 |
|---|---|
| 처음부터 RAG pipeline 구현하기 - (2) (1) | 2024.12.17 |
| Hugging Face를 이용한 Transformer 모델 활용 실습 (6) | 2024.12.11 |
| KSS 데이터셋으로 TorToise-tts 한국어 Fine-tuning (6) | 2024.11.05 |
| Word representation & Feed Forward Neural Network Language Model(NNLM) (1) | 2024.03.07 |
